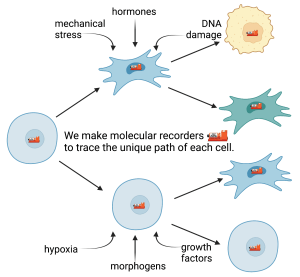
During the development of multicellular organisms, and in aberrant developmental processes such as cancer, cells undergo a series of stresses, gene expression changes, and other transient events, some of which determine their ultimate fates. The historical events preceding most cell fates are unknown. Even less is known about which historical events cause a cell fate and which merely accompany it. DNA recorders are synthetic biology tools that enable cells to make a durable DNA record of ephemeral events in their history, in principle enabling biologists to correlate any type of cellular event to long term changes in the cell’s phenotype. Moreover, by recording the relative timings of the events and the fate changes, DNA recorders should make it possible to form better hypotheses about which events cause which fate changes. Our lab’s goal is to realize this promise by engineering better DNA recorders. One application of particular interest to us is long-term genotoxic stress and its consequences for cell fate. As the lab grows and our technology matures, our tools for recording cell history will also be adapted to perturb cell fate in response to specific histories.
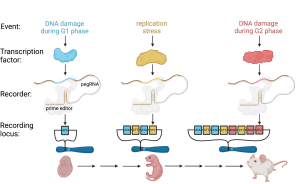 The enabling technology for our DNA recorders, which we call peCHYRON, was developed during Theresa’s postdoc. So far, we have a prototype that works in tissue culture cells. The above figure shows how it could be used to study DNA damage during devleopment. peCHYRON operates by expressing prime editor, a Cas9 nickase-reverse transcriptase (RT) fusion protein, and a series of prime editor guide RNAs (pegRNAs). pegRNAs combine a single-guide RNA (sgRNA), to direct the Cas9 component to the target site, with a primer binding sequence and RT template, to specify the mutation made. The sgRNA component of the pegRNAs directs prime editor to a “recording locus,” which can in principle be any genomic site. The RT template portion of each pegRNA defines an insertion mutation with two parts: a “signature sequence” and a “propagation sequence.” The signature sequence (blue, yellow, or red) is different in each pegRNA and is permanently incorporated into the recording locus; all recorded signature sequences constitute the record. The propagation sequence (brown) is the same in all the pegRNAs; it directs the destruction of the target site upstream of the signature sequence and the creation of a new target site downstream, which can be targeted in the next round. As development progresses, different transcriptional activators present in the cell’s nucleus direct the expression of specific pegRNAs. In this example, the transcription factors whose activity is recorded are active in different parts of the DNA damage response. However, additional transcription factors could be added, such as those active at known times during development, and in principle any transcriptional event could be recorded.
The enabling technology for our DNA recorders, which we call peCHYRON, was developed during Theresa’s postdoc. So far, we have a prototype that works in tissue culture cells. The above figure shows how it could be used to study DNA damage during devleopment. peCHYRON operates by expressing prime editor, a Cas9 nickase-reverse transcriptase (RT) fusion protein, and a series of prime editor guide RNAs (pegRNAs). pegRNAs combine a single-guide RNA (sgRNA), to direct the Cas9 component to the target site, with a primer binding sequence and RT template, to specify the mutation made. The sgRNA component of the pegRNAs directs prime editor to a “recording locus,” which can in principle be any genomic site. The RT template portion of each pegRNA defines an insertion mutation with two parts: a “signature sequence” and a “propagation sequence.” The signature sequence (blue, yellow, or red) is different in each pegRNA and is permanently incorporated into the recording locus; all recorded signature sequences constitute the record. The propagation sequence (brown) is the same in all the pegRNAs; it directs the destruction of the target site upstream of the signature sequence and the creation of a new target site downstream, which can be targeted in the next round. As development progresses, different transcriptional activators present in the cell’s nucleus direct the expression of specific pegRNAs. In this example, the transcription factors whose activity is recorded are active in different parts of the DNA damage response. However, additional transcription factors could be added, such as those active at known times during development, and in principle any transcriptional event could be recorded.
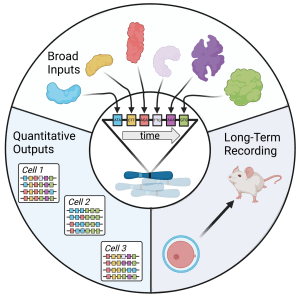
Ultimately, our DNA recorders will need to be able to record a broad range of inputs, quantitatively, over organismal lifetimes.
Broad inputs. Transcription levels of specific genes can be recorded by inserting pegRNA expression cassettes into introns, and activities of specific transcription factors can be recorded using well-validated promoters that bind specifically to that factor. By performing these types of analyses on related pathways in parallel in the same cell, webs of transcriptional regulation over time would be revealed. Moreover, as ever more ways to control the activities of guide RNAs are invented, it is possible to envision the recording of non-transcriptional events, such as chemical exposures or levels of non-coding RNAs.
Quantitative outputs. Ideally, if multiple reporter pegRNAs are expressed at the same time, their recording at multiple genomic sites will be proportional to their expression. Achieving this goal will require tuning the expression levels of prime editor and the half-lives of recorder pegRNAs. In addition, the multiple recorders in each single cell will need to be aligned to each other at the end of the experiment, so that it is known which events happened at the same time. For this purpose, we could, for example, express both constitutive “internal timekeeper” pegRNAs that record at a constant rate, and “external timekeeper” pegRNAs that are strongly induced by a drug given to the animal, such as doxycycline.
Long-term recording. In order to faithfully record events over the lifetime of an experimental animal, I will need to make recording components that aren’t significantly silenced in vivo and show that they do not themselves perturb the developmental processes they aim to study.